Datasets & Data analysis
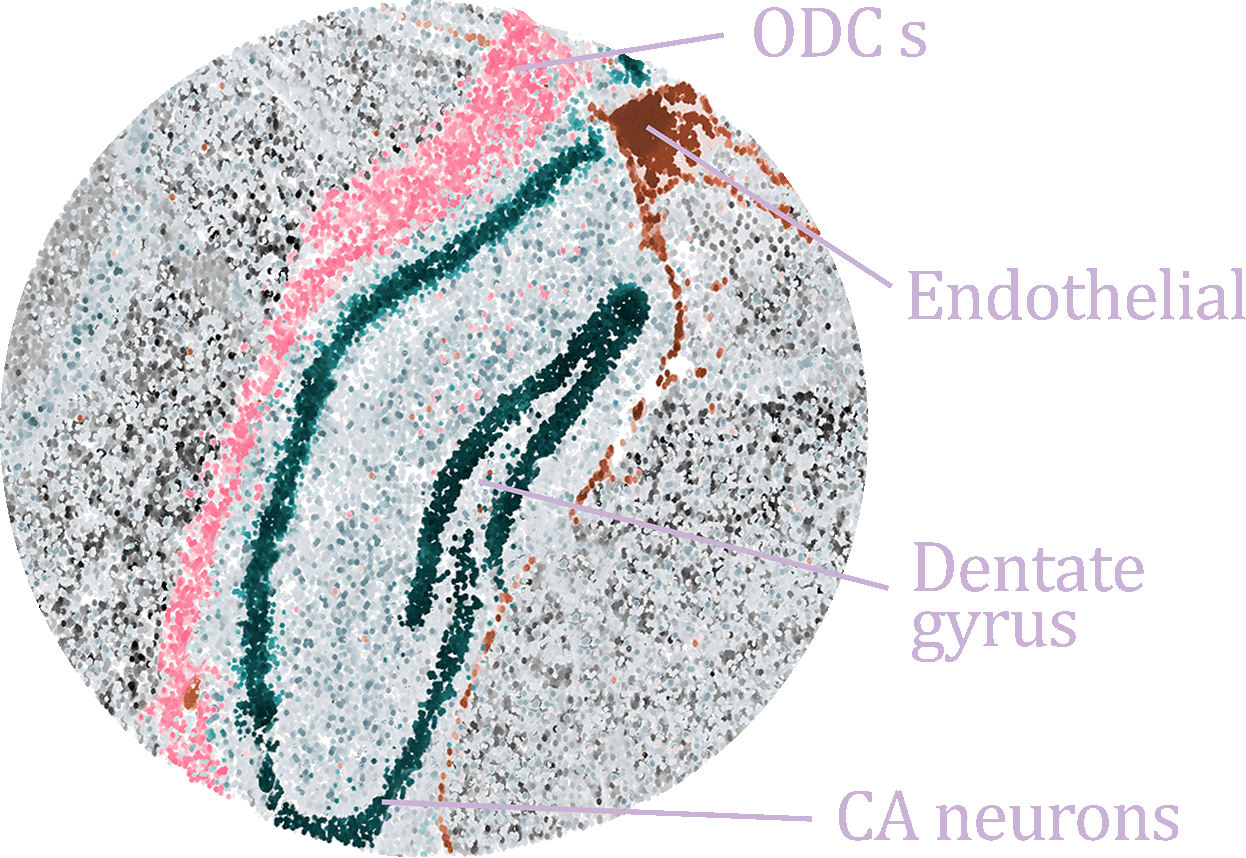
Hippocampus Cell Atlas (HCA)
A comprehensive single-cell atlas integrating over 420,000 cells across 33 distinct cell types from multiple public datasets
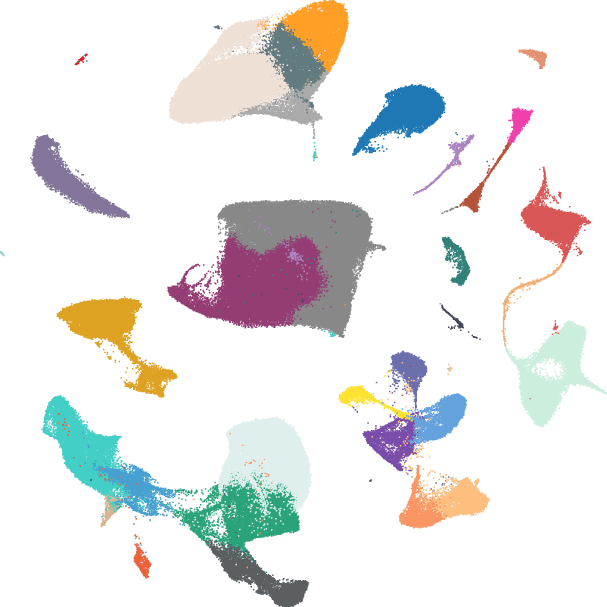
- Interactive visualization of hippocampal cell types
- Detailed gene expression patterns
- Advanced co-expression analysis tools, with user-provided gene set overlay capabilities
Post- hypoxia and ischemia
An atlas of >27,000 nuclei profiled at critical early (3-hour) timepoint in sham and post-hypoxia-ischemia.
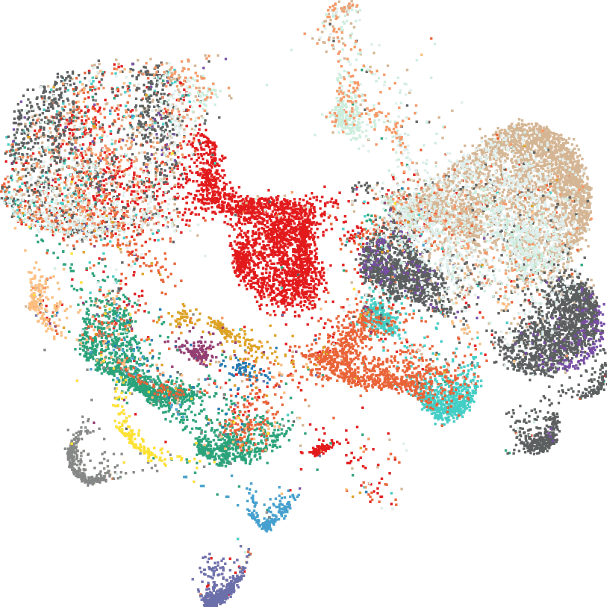
- Cell-type specific vulnerability analysis
- Transcriptional changes during early response
- Interactive comparison across conditions
- Advanced co-expression analysis tools, with user-provided gene set overlay capabilities
Data analysis
sc/nRNA-seq analysis of user provided data
- Discover what cell types your transcriptomics and spatial data correspond to. Extract cluster-specific differentially expressed genes from your data and overlay on the atlases to see corresponding cell types.
- Overlay disease-associated genes to identify potential involvement in specific hippocampal cell types or pathological conditions.
- Automatically assign cell types for Hippocampus through R or Python with ScType using our ML-based identified hippocampus markers.